🚀 Try Me – One Click, Done Quick!
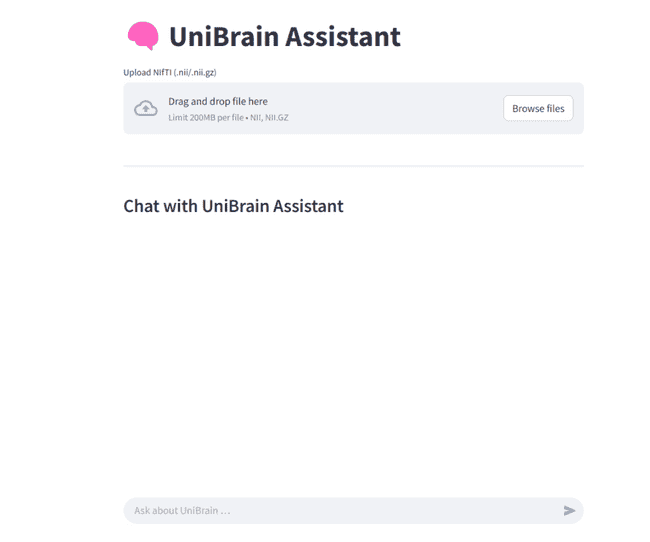
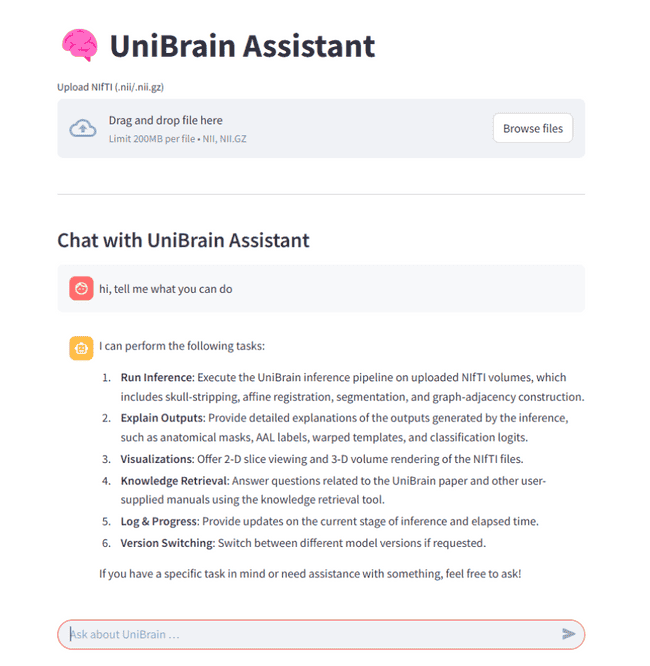
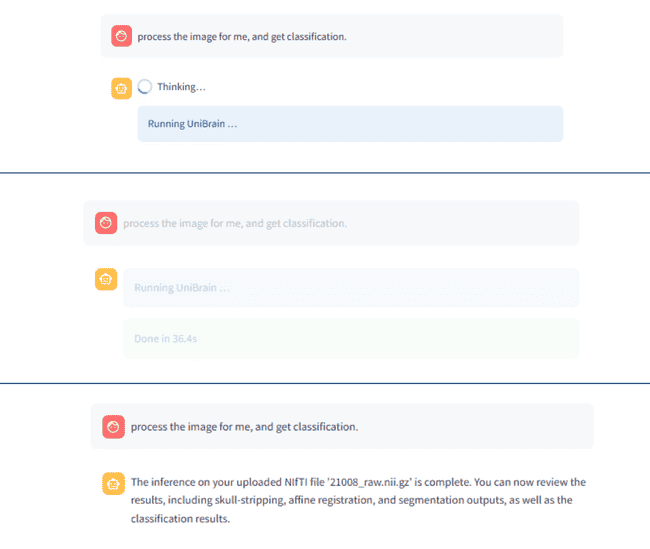
You can drop in a NIfTI file, watch every preprocessing step unfold in real time, explore the resulting connectome interactively, and ask questions in plain English or any natural language — all without leaving your web browser.
It pairs Streamlit’s reactive UI with LangChain’s tool-calling so you can see, tweak, and interrogate each stage of the pipeline:
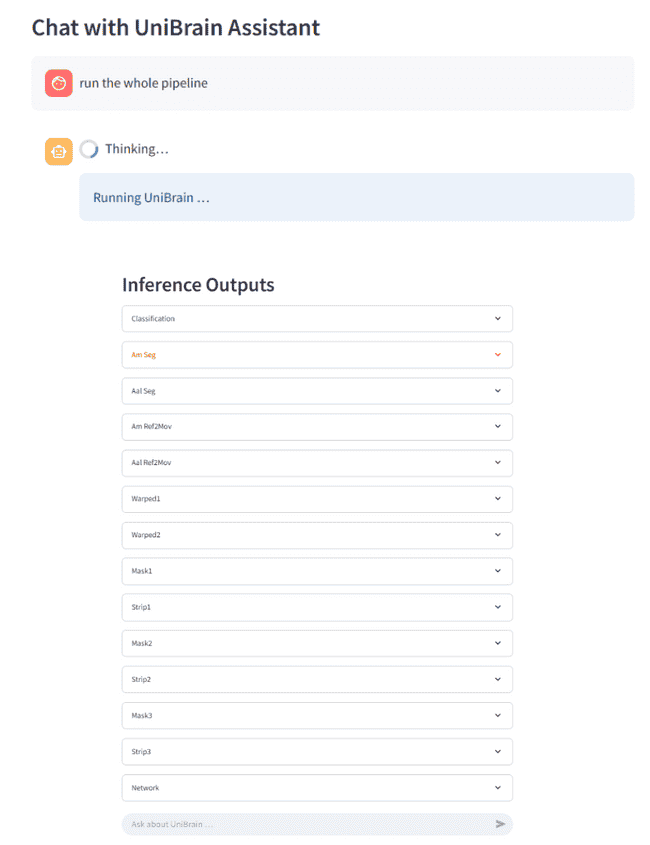
- skull-stripping → affine registration → tissue segmentation → AAL parcellation → graph construction → disease classification
- fully interactive: 2D slice viewer, 3D Plotly volume, heat-map / graph visualizations, one-click downloads
- pipeline orchestration by natural-language – e.g.
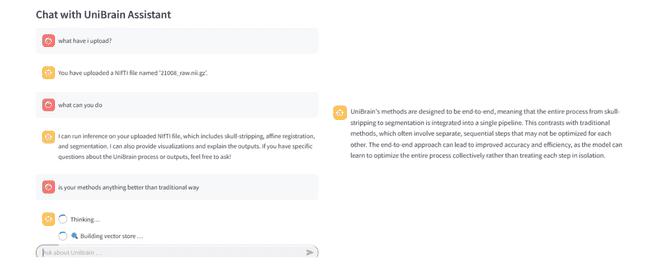
run the pipeline without segmentation,enable network - RAG-powered Q & A over both your outputs and the UniBrain paper itself
⏳ We are working hard to enhance the tool, and a new version will be released soon.
🚀 Quick Start (Self-Deployment)
git clone https://github.com/soz223/UniBrainAssistant.git
cd UniBrainAssistant
python3 -m venv .venv && source .venv/bin/activate # optional
pip install -r requirements.txt
export OPENAI_API_KEY="sk-..." # for LLM chat & RAG
streamlit run app.pyadd something
Open http://localhost:8501 → upload a NIfTI (or DICOM) → pick processing steps → Run. Then interact with your scan:
❯ skip segmentation # run without the Segmentation step
❯ what does a high dice score mean?
❯ show only the network analysis stage🔑 Environment Variables
| Var | Purpose |
|---|---|
OPENAI_API_KEY |
Required for chat, command-parser LLM, and RAG |
IMG_SIZE |
(optional) override default 96³ voxel input size |
ATLAS |
(optional) specify brain atlas (e.g., “Desikan”, “AAL”) |
📦 Core Dependencies
- UI & Workflow:
streamlit ≥1.32 - Imaging & ML:
torch,numpy,nibabel,SimpleITK,plotly,networkx - LLM & RAG:
langchain,langchain-openai,faiss-cpu,openai (≥1.0)
See requirements.txt for exact version pins.
🤖 Command Grammar (for reference)
| Intent | Examples (case-insensitive) |
|---|---|
| Skip step | skip segmentation, no parcellation, without network |
| Enable step | enable analysis, turn on registration, enable classification |
| Reset | reset steps, reset pipeline |
| General chat | anything else → routed to the UniBrain chat assistant |
Processing flow:
- Regex fast-path
- if unresolved → GPT-4o-mini (
CMD_SYS_PROMPT) → JSON command
🌐 Try the Demo Website
-
Visit the live demo at Ongoing Website
-
Enter your
OPENAI_API_KEYin the sidebar API key field -
Upload a
.niior.nii.gzfile -
Use the sidebar menu to enable/disable steps, then click Run
-
Chat with your data right in the browser:
❯ What can you do? ❯ Run the whole pipeline on the uploaded image ❯ Skip segmentation and do the whole preprocessing ❯ How does parcellation work?